Global Journal of Infectious Diseases and Clinical Research
The importance of the ecology of infectious diseases in the context of Chytridiomycosis and COVID-19
Koichi Goka*
Cite this as
Goka K (2023) The importance of the ecology of infectious diseases in the context of Chytridiomycosis and COVID-19. Glob J Infect Dis Clin Res 9(1): 001-008. DOI: 10.17352/2455-5363.000054Copyright
© 2023 Goka K. This is an open-access article distributed under the terms of the Creative Commons Attribution License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original author and source are credited.Pathogenic microorganisms and viruses are components of ecosystems. They have constructed endemic interrelationships with specific host species throughout the history of coevolution The spillover of pathogens from natural habitats into other areas causes encounters between the pathogens and new hosts that have never evolved immunity or resistance. The result is a rapid spread of “Emerging Infectious Disease” (EID).
During the acceleration of globalization, humans and societies have come to be the targets of infectious diseases caused by pathogens that have emerged from natural habitats. In the past few years, the newest EID, SARS-CoV-2, has spread throughout the world and has caused serious harm to human health and welfare.
With the growing social concern about the risks of the EID pandemic, there has been discussion that the destruction of biodiversity and environmental changes are closely related to the EID pandemic. There is a need to rethink the importance of conserving biodiversity if humans are to control the risk of pathogenic viruses and live in harmony with them.
Introduction
My own specialty is conservation ecology and my work at the National Institute for Environmental Studies involves countermeasures to the risk of invasive alien species. It is a bit of a stretch for an ecologist to become involved in a field with such serious implications for public health as infectious disease, but I think that biodiversity and various environmental issues are closely associated with infectious disease pandemics and I believe that cross-disciplinary discussions and research are needed to address the problems caused by infectious diseases.
In this paper, I review the results of my own work as a conservation ecologist on the Chytridiomycosis pandemic. I discuss the diversity and endemism of pathogens and the need to understand the evolutionary and ecological aspects of pathogens in order to manage the risks associated with infectious diseases.
I surveyed the literature on the origin of COVID-19 and its impact on human societies and ecosystems using Google Scholar, CiNii and J-STAGE. I carried out an evolutionary, ecological analysis of the COVID-19 pandemic over the past three years with the expectation that the analysis would reveal the importance of biodiversity conservation and global environmental management in the fight against emerging infectious diseases in the future.
Emerging infectious diseases of amphibians: chytridiomycosis
I first became involved in infectious disease issues because of my interest in a fungal disease specific to amphibians called chytridiomycosis. Since the 1990s, the fungus has spread throughout the world and has caused a biodiversity crisis by threatening endangered amphibians with extinction [1-3]. In 2006, the disease was confirmed in Japan in an Argentine horned frog, Ceratophrys ornata, that had been kept as an exotic pet [4]. This discovery led to fears that the chytridiomycosis invasion would lead to the extinction of Japanese amphibians.
Our research team investigated the status of infection of amphibians throughout Japan and overseas, and we collected and analyzed samples of chytridiomycosis DNA. Our analyses indicated that the endemic Japanese giant salamander, Andrias japonicus, and the Okinawa sward tail newt, Cynops ensicauda popei, were likely the original hosts of the fungus and further experiments showed that Japanese frogs had developed resistance to the fungus.
We concluded from our analysis of these data that the fungus had originated in Asia, including Japan, and that native amphibians in Japan had acquired resistance to the fungus through a long period of coevolution with the fungus [5,6]. Subsequent global DNA surveys of the chytrid fungus by European and American research groups have also suggested that the fungus originated in Asia [7,8]. The fungus was likely introduced from Asia into various countries around the world during the international transportation of various amphibians for food and pet markets. The beginning of international transfer of the fungus may date back to the beginning of Japanese immigration to Brazil around 1900 [6,9,10] (Figure 1).
Serious impacts caused by the chytrid fungus have been concentrated mainly in rare amphibian populations in high-altitude jungle areas of Latin America and Oceania. Excessive tourism in natural forest areas (e.g., ecotourism) has been identified as a cause of the chytrid fungus’ entering deep jungle areas.
The results of a series of studies of the chytrid fungus have provided important insights into the ecological aspects of infectious diseases. Pathogens such as fungi and viruses are diverse. Some are endemic to particular areas and/or originated in particular habitats. During their coevolution with other wildlife in local ecosystems, some micro-organisms establish symbiotic relationships with wildlife. When humans encroach on natural ecosystems and export pathogens that have been isolated there, they can spread the pathogen to other habitats and cause an infectious explosion of the pathogen among animals in other ecosystems that are not resistant to the pathogen.
This process is consistent with the mechanisms that have caused the infectious disease outbreaks that currently pose a threat to human societies. The case of the infection of frogs by the chytrid fungus suggests the need to recognize the importance of biodiversity and ecosystem conservation to curb infectious disease outbreaks in human societies.
Biodiversity disturbance and emerging infectious diseases
The world of wildlife is home to a variety of pathogenic microorganisms, including fungi, bacteria and viruses. From the human perspective, pathogens may seem like a nuisance, but as members of the biota, they have evolved among wildlife populations since ancient times [11-15] and are thought to have played an important ecosystem function.
In other words, pathogens are thought to function as internal natural enemies in nature by parasitizing or infecting populations of organisms to weed out individuals with weak resistance or immunity and adjust the population size. The result is the maintenance of the balance of the ecosystem[16-18].
For example, when the red tide plankton Heterocapsa circularisquama blooms in unusually high numbers in the ocean, the “natural enemy virus” HcRNAV infects H. circularisquama and reduces its density [19]. Even in the world of spider mites, there are specific parasitic fungi that spread infection and suppress population density when spider mite populations become dense [20,21].
In addition, interactions between pathogens and hosts, such as the arms race coevolution of virulence and immunity, have been important evolutionary drivers of diversity in both pathogens and host organisms [22-28].
Since the 1970s, highly lethal viral infectious diseases such as AIDS (Acquired Immune Deficiency Syndrome), Ebola, West Nile Fever, SARS (Severe Acute Respiratory Syndrome), MERS (Middle East Respiratory Syndrome) and new strains of influenza have been emerging around the world and causing serious health problems for human societies. Many of the pathogens associated with these emerging infectious diseases are considered to have originated from wild animals [29-32]. It is believed that these epidemics are due to the fact that humans have encroached deeper into the world of wildlife [33-37].
Johnson, et al. [38] carried out a statistical analysis of global data on zoonotic and related viruses transmitted by various groups of animals and found that the status of domesticated species has the largest influence on the number of mammalian viruses shared with humans. They found that there were eight times more zoonotic viruses in a given species of domesticated mammal than in wild mammalian species. Their network analysis of virus-sharing relationships among wild and domestic animals indicates that domestic animals share viruses that cause infectious diseases with various wild animals. These results mean that species of domestic animals have become an interface between the natural world and human society in the transmission of viruses as areas of domestic animal husbandry have encroached into wildlife habitats.
The network analysis also indicated that primate, rodent and bat species appear to harbor zoonotic viruses that are not well connected to domesticated species and other wild animals. The implication is that these species share zoonotic viruses directly with humans, without amplification by domesticated hosts to facilitate viral sharing among species in other orders. For example, it has been noted that spillovers of HIV, which originated in monkeys and the SARS virus, which originated in bats, were caused by close contact between wild animals and humans through hunting and trafficking [39-42].
Johnson, et al. [38] found that threatened species whose population size has been reduced due to exploitation have over twice as many zoonotic viruses as threatened species listed for other reasons. This discovery points to the need for strict measures to control infection of (i) those involved in hunting and trading, (ii) researchers and activists involved in the conservation and protection of threatened animals and (iii) people involved in nature tourism industries such as ecotourism, who observe these animals (including tourists).
For example, most biologists are at a loss to answer the question of whether they have ever paid attention to the microorganisms that accompany the soles of their shoes during overseas field research. In many cases, biologists have captured animals with their bare hands. In fact, there have recently been cases in Japan where such images have been sold in the popular media. However, unintentional direct or indirect contact with wild animals poses not only the risk of pathogens being transmitted from the animals to humans but also the risk of transmission of exotic pathogens from the humans to the wild animals [43-46]. Researchers need to be aware of their own risk of infectious disease transmission.
Ecological implications of COVID-19: origins and spillovers
The novel coronavirus SARS-CoV-2, which has continued to cause serious human health problems since the first infection was reported in China in late 2019, is also presumed to be of wild animal origin.
SARS-CoV-2-related viruses have been reported from various Rhinolophus bat species inhabiting Asia, including China [47], Cambodia [48], Thailand [49], and Japan [50]. The lineage RaTG13, which is carried by Rhinolophus ferrumequinum captured in Yunnan, China, is considered to be most closely related to the novel coronavirus at the whole genome level [51]. Because the coronavirus harbored by pangolins also exhibits RNA sequences homologous to SARS-CoV-2, it has been suggested that this animal is also involved in the origin of the new infectious coronavirus [52-54].
Temmam, et al. [55] investigated coronaviruses infecting Rhinolophus bats in Southeast Asia by focusing on the homology of the spike protein sequence, which plays an important role in infecting human cells. They found several viral strains whose receptor binding domain (RBD) sequences of spike protein more closely resembled those of SARS-CoV-2 than those of RaTG13.
These results suggest that a group of viruses that are precursors of SARS-CoV-2 have been circulating and evolving in wild animal populations such as bats and pangolins in the Asian region. In recent years, deforestation for the purpose of road development, drilling for mineral deposits and expansion of agricultural land has been rapidly progressing in China and Southeast Asia. It is speculated that the human disturbance of the wildlife-virus symbiosis zone in these areas has given SARS-CoV-2, which happened to have evolved into a human form, a chance to spill over into humans.
Ecological implications in COVID-19: infection spread and evolution
COVID-19, which spilled over from the wildlife world into human society, spread to all regions of the world in just a few months after the first cases were identified. The speed of the spread of COVID-19 suggests the virus’ strong infectivity as well as its adaptability and environmental tolerance, which have allowed it to spread into any environment, regardless of climate or altitude.
The continued evolution of the virus in humans through mutation and genetic recombination [56] has resulted in mutant strains with further enhanced infectivity, virulence and resistance to immunity [57]. Since the global epidemic began, the number of new cases of this virus has fluctuated from country to country and region to region, but on a global level, the figure has always been several hundred thousand or more per day and the infection is always spreading somewhere in the world [58].
If this situation is viewed from a population ecology perspective, regional populations of viruses form a metapopulation structure in which they are connected to each other through gene flow. Each population increases or decreases in number more or less independently but the risk of extinction is very low thanks to the connections between populations. This metapopulation structure is thought to support a large population of genetically very diverse viruses and to sustain the evolution and spread of new variants. Until effective therapeutic agents are available worldwide, the effective population size of the virus must first be kept low to prevent the emergence of new mutant strains and to protect the medical community. It is important for each country to keep a close watch on the virus mutation and take adaptive measures to prevent infection.
Many cases of SARS-CoV-2 being transmitted from humans to captive and wild animals have been reported, and in 2022, a viral strain that evolved specifically within white-tailed deer in Canada was reported to infect humans [59]. This suggests that SARS-CoV-2 can evolve repeatedly in wild animals and produce a spill-back that can infect humans again. Surveillance of SARS-CoV-2 in wild animal species will therefore be an important issue in the future.
Sociological lessons from COVID-19: independence and equality required of states and the international community
The reason for the rapid global spread of COVID-19, arguably the fastest in the history of infectious diseases, can be attributed not only to the infectiousness of the virus itself but also to an overly-growing global economy. Taking advantage of the high-speed human flow network and over-tourism around the world, the virus spread rapidly to the remote areas of the Amazon and to localized areas in the North and South. (Of personal interest was the fact that in 2020, despite reports of the spread of SARS-CoV-2 to ethnic minorities in Brazil, Iwate Prefecture in Japan was defending itself with zero cases of infection).
Because the entire world is so dependent on the global economy, the stagnation of human flows and logistics after the virus spread resulted in tremendous damage to the economies of countries around the world [60]. Lenzen, et al. [61] analyzed supply chain losses in each country during the first wave of the COVID-19 pandemic and found that supply chain losses were particularly large in China, the United States, and Europe. The indication is that these countries and regions are the mainstays of the international economy.
In a global economy where efficiency is a priority, the production of many products, both agricultural and industrial, is concentrated in regions where manufacturing costs can be kept low. In particular, the entire world is dependent on the global supply chain for most medical supplies. When the global supply chain was disrupted by COVID-19 and as demand rapidly increased, the entire world, therefore, ran out of masks and ventilators, and treatment could not keep up with the rapid increase in the number of infected people. The result was a risk of medical collapse [62]. It can be said that the large damage caused by COVID-19 was brought about by the over-globalization of human activities.
Furthermore, the economic and medical crisis has driven many countries to nationalism, which has accelerated the prioritization of national and individual economies. That prioritization has resulted in the stockpiling of natural resources and vaccines as well as the monopolization of profits. The result has been a further increase in disparities and inequalities among countries and their citizens [63-65]. The relentless attack of COVID-19 on the socially vulnerable, including those who lacked access to medical care because of poverty or were forced to work in jobs with a high risk of infection, spurred the spread of the disease [66-70].
Ecological implications of COVID-19: emerging infectious diseases and global environmental issues
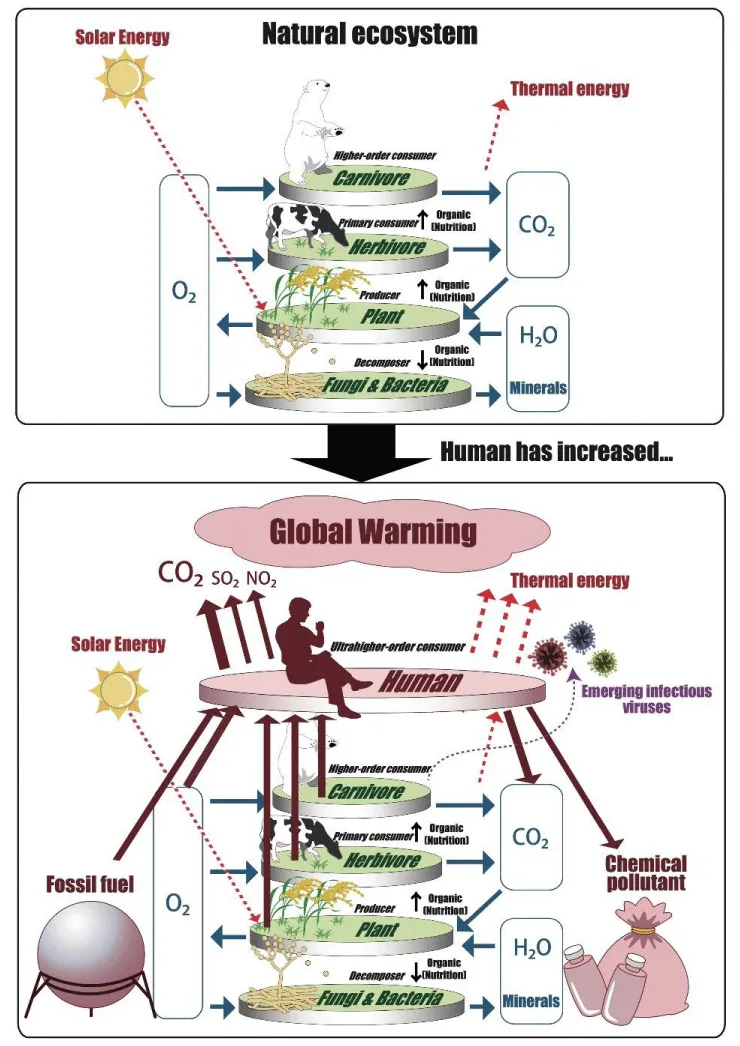
From a macroscopic perspective of the global ecosystem, the COVID-19 pandemic can be viewed as a natural phenomenon that was bound to happen. In a natural ecosystem, the only energy supplied by the outside world is solar energy. Plants use this solar energy to produce organic matter through photosynthesis and animals use the organic matter as a source of energy for vital activities. When plants and animals die, they become detritus, which is decomposed by detritivores into inorganic substances that again become raw materials for photosynthesis. In this way, natural ecosystems have been maintained as fully cyclical systems (top panel in Figure 2).
The arrival of humans has upset the balance and sustainability of the global ecosystem. Humans are apex predators, and the human population now numbers 8 billion. Humans exploit all resources from the natural ecosystem while continuing to rely on fossil fuels such as oil for energy and the production of materials. They release massive amounts of waste and greenhouse gases into the natural world (bottom panel in Figure 2).
When animal populations become so numerous that they place a heavy burden on the ecosystem, natural enemies evolve within resilient ecosystems. At the present time, the natural enemies of humans, who are causing the environmental burden, are precisely the viruses that cause emerging infectious diseases (bottom panel in Figure 2).
Human populations, with their enormous energy reserves, are easy prey for pathogenic viruses, and there is no way to avoid infection. The emergence of infectious diseases is a natural consequence of the natural order of things and the emergence of COVID-19 is a poster child for the era of human-caused environmental destruction.
Conclusion
The occurrence of two very different infectious disease pandemics, Chytridiomychosis and COVID-19, has led to a common theory: pathogens have a native habitat and a natural host. Humans have entered that habitat and/or displaced the natural host. The result has been pathogen spillover.
Conserving biodiversity and maintaining a precise distance between humans and wildlife are considered critical to preventing infectious disease pandemics. The COVID-19 pandemic, in particular, exposed the fragility of crisis management that was brought about by the global economy. Because the COVID-19 pandemic also contributed to economic inequality and discrimination in human societies, it increased the scale of casualties. This global epidemic showed that inequality and poverty in human societies are major obstacles to disaster risk management.
The phenomenon of emerging infectious disease viruses evolving and attacking humans as a natural enemy must be viewed ecologically as a mechanism of ecological resilience. To control the risk of infectious diseases and build a safe and secure society in the future, humans must aim for social equity and peace and at the same time, we must transform to a nature-friendly way of life that enables the sustainable use of natural resources.
Limitations
This review is only an attempt by the author, a conservation ecologist, to present ecological suggestions for countermeasures against emerging infectious disease risks, including COVID-19, based on extant data. The attempt will require analysis and validation from the professional perspectives of virology, pathology and epidemiology.
It is my hope that this review will promote ecological- and environmental-science studies of the COVID-19 pandemic and that the results from these new studies will contribute to the future development of ways to control emerging infectious diseases.
- Berger L, Speare R, Daszak P, Green DE, Cunningham AA, Goggin CL, Slocombe R, Ragan MA, Hyatt AD, McDonald KR, Hines HB, Lips KR, Marantelli G, Parkes H. Chytridiomycosis causes amphibian mortality associated with population declines in the rain forests of Australia and Central America. Proc Natl Acad Sci U S A. 1998 Jul 21;95(15):9031-6. doi: 10.1073/pnas.95.15.9031. PMID: 9671799; PMCID: PMC21197.
- Weldon C, du Preez LH, Hyatt AD, Muller R, Spears R. Origin of the amphibian chytrid fungus. Emerg Infect Dis. 2004 Dec;10(12):2100-5. doi: 10.3201/eid1012.030804. PMID: 15663845; PMCID: PMC3323396.
- Lips KR, Brem F, Brenes R, Reeve JD, Alford RA, Voyles J, Carey C, Livo L, Pessier AP, Collins JP. Emerging infectious disease and the loss of biodiversity in a Neotropical amphibian community. Proc Natl Acad Sci U S A. 2006 Feb 28;103(9):3165-70. doi: 10.1073/pnas.0506889103. Epub 2006 Feb 15. PMID: 16481617; PMCID: PMC1413869.
- Une Y, Kadekaru S, Tamukai K, Goka K, Kuroki T. First report of spontaneous chytridiomycosis in frogs in Asia. Dis Aquat Organ. 2008 Nov 20;82(2):157-60. doi: 10.3354/dao02006. PMID: 19149379.
- Goka K, Yokoyama J, Une Y, Kuroki T, Suzuki K, Nakahara M, Kobayashi A, Inaba S, Mizutani T, Hyatt AD. Amphibian chytridiomycosis in Japan: distribution, haplotypes and possible route of entry into Japan. Mol Ecol. 2009 Dec;18(23):4757-74. doi: 10.1111/j.1365-294X.2009.04384.x. Epub 2009 Oct 13. PMID: 19840263.
- Goka K, Yokoyama J, Tominaga A. Distribution and Genetic Diversity of the Amphibian Chytrid in Japan. J Fungi (Basel). 2021 Jun 29;7(7):522. doi: 10.3390/jof7070522. PMID: 34210103; PMCID: PMC8307550.
- O'Hanlon SJ, Rieux A, Farrer RA, Rosa GM, Waldman B, Bataille A, Kosch TA, Murray KA, Brankovics B, Fumagalli M, Martin MD, Wales N, Alvarado-Rybak M, Bates KA, Berger L, Böll S, Brookes L, Clare F, Courtois EA, Cunningham AA, Doherty-Bone TM, Ghosh P, Gower DJ, Hintz WE, Höglund J, Jenkinson TS, Lin CF, Laurila A, Loyau A, Martel A, Meurling S, Miaud C, Minting P, Pasmans F, Schmeller DS, Schmidt BR, Shelton JMG, Skerratt LF, Smith F, Soto-Azat C, Spagnoletti M, Tessa G, Toledo LF, Valenzuela-Sánchez A, Verster R, Vörös J, Webb RJ, Wierzbicki C, Wombwell E, Zamudio KR, Aanensen DM, James TY, Gilbert MTP, Weldon C, Bosch J, Balloux F, Garner TWJ, Fisher MC. Recent Asian origin of chytrid fungi causing global amphibian declines. Science. 2018 May 11;360(6389):621-627. doi: 10.1126/science.aar1965. PMID: 29748278; PMCID: PMC6311102.
- Byrne AQ, Vredenburg VT, Martel A, Pasmans F, Bell RC, Blackburn DC, Bletz MC, Bosch J, Briggs CJ, Brown RM, Catenazzi A, Familiar López M, Figueroa-Valenzuela R, Ghose SL, Jaeger JR, Jani AJ, Jirku M, Knapp RA, Muñoz A, Portik DM, Richards-Zawacki CL, Rockney H, Rovito SM, Stark T, Sulaeman H, Tao NT, Voyles J, Waddle AW, Yuan Z, Rosenblum EB. Cryptic diversity of a widespread global pathogen reveals expanded threats to amphibian conservation. Proc Natl Acad Sci U S A. 2019 Oct 8;116(41):20382-20387. doi: 10.1073/pnas.1908289116. Epub 2019 Sep 23. PMID: 31548391; PMCID: PMC6789904.
- Rodriguez D, Becker CG, Pupin NC, Haddad CF, Zamudio KR. Long-term endemism of two highly divergent lineages of the amphibian-killing fungus in the Atlantic Forest of Brazil. Mol Ecol. 2014 Feb;23(4):774-87. doi: 10.1111/mec.12615. PMID: 24471406.
- Fisher MC, Garner TWJ. Chytrid fungi and global amphibian declines. Nat Rev Microbiol. 2020 Jun;18(6):332-343. doi: 10.1038/s41579-020-0335-x. Epub 2020 Feb 25. PMID: 32099078.
- Suttle CA. Viruses: unlocking the greatest biodiversity on Earth. Genome. 2013 Oct;56(10):542-4. doi: 10.1139/gen-2013-0152. PMID: 24237332.
- Obbard DJ, Dudas G. The genetics of host-virus coevolution in invertebrates. Curr Opin Virol. 2014 Oct;8:73-8. doi: 10.1016/j.coviro.2014.07.002. Epub 2014 Jul 24. PMID: 25063907; PMCID: PMC4199324.
- Polinar G. Evolutionary History of Terrestrial Pathogens and Endoparasites as Revealed in Fossils and Subfossils. Advances in Biology. 2014. https://doi.org/10.1155/2014/181353
- Larsen BB, Miller EC, Rhodes MK, Wiens JJ. Inordinate fondness multiplied and redistributed: the number of species on Earth and the new pie of life. Q Rev Biol. 2017; 92(3):229–265. https://doi.org/10.1086/693564
- Kaján GL, Doszpoly A, Tarján ZL, Vidovszky MZ, Papp T. Virus-Host Coevolution with a Focus on Animal and Human DNA Viruses. J Mol Evol. 2020 Jan;88(1):41-56. doi: 10.1007/s00239-019-09913-4. Epub 2019 Oct 10. PMID: 31599342; PMCID: PMC6943099.
- Rohwer F, Prangishvili D, Lindell D. Roles of viruses in the environment. Environ Microbiol. 2009 Nov;11(11):2771-4. doi: 10.1111/j.1462-2920.2009.02101.x. PMID: 19878268.
- Fischhoff IR, Huang T, Hamilton SK, Han BA, LaDeau SL, Ostfeld RS, Rosi EJ, Solomon CT. Parasite and pathogen effects on ecosystem processes: A quantitative review. Ecosphere. 2020; 11(5):e03057. https://doi.org/10.1002/ecs2. 3057
- Paseka RE, White LA, Van de Waal DB, Strauss AT, González AL, Everett RA, Peace A, Seabloom EW, Frenken T, Borer ET. Disease-mediated ecosystem services: Pathogens, plants, and people. Trends Ecol Evol. 2020 Aug;35(8):731-743. doi: 10.1016/j.tree.2020.04.003. Epub 2020 Jun 15. PMID: 32553885.
- Nagasaki K, Takao Y, Shirai Y, Mizumoto H, Tomaru Y. [Molecular ecology of microalgal viruses]. Uirusu. 2005 Jun;55(1):127-32. Japanese. doi: 10.2222/jsv.55.127. PMID: 16308539.
- Elliot SL, de Moraes GJ, Delalibera I Jr, da Silva CA, Tamai MA, Mumford JD. Potential of the mite-pathogenic fungus Neozygites floridana (Entomophthorales: Neozygitaceae) for control of the cassava green mite Mononychellus tanajoa (Acari: Tetranychidae). Bull Entomol Res. 2000 Jun;90(3):191-200. doi: 10.1017/s0007485300000316. PMID: 10996860.
- Ishikawa I. Occurrence of Neozygites sp. (Zygomycetes: Entomophthorales) attacking the Kanzawa spider mite, Tetranychus kanzawai (Kishida) in a tea field. Tea research journal. 2010; 109: 65-72.
- Ebert D, Hamilton WD. Sex against virulence: the coevolution of parasitic diseases. Trends Ecol Evol. 1996 Feb;11(2):79-82. doi: 10.1016/0169-5347(96)81047-0. PMID: 21237766.
- Dybdahl MF, Lively CM. HOST-PARASITE COEVOLUTION: EVIDENCE FOR RARE ADVANTAGE AND TIME-LAGGED SELECTION IN A NATURAL POPULATION. Evolution. 1998 Aug;52(4):1057-1066. doi: 10.1111/j.1558-5646.1998.tb01833.x. PMID: 28565221.
- Koonin EV, Dolja VV. A virocentric perspective on the evolution of life. Curr Opin Virol. 2013 Oct;3(5):546-57. doi: 10.1016/j.coviro.2013.06.008. Epub 2013 Jul 12. PMID: 23850169; PMCID: PMC4326007.
- Koskella B. Resistance gained, resistance lost: An explanation for host-parasite coexistence. PLoS Biol. 2018 Sep 24;16(9):e3000013. doi: 10.1371/journal.pbio.3000013. PMID: 30248103; PMCID: PMC6171958.
- Duxbury EM, Day JP, Maria Vespasiani D, Thüringer Y, Tolosana I, Smith SC, Tagliaferri L, Kamacioglu A, Lindsley I, Love L, Unckless RL, Jiggins FM, Longdon B. Host-pathogen coevolution increases genetic variation in susceptibility to infection. Elife. 2019 Apr 30;8:e46440. doi: 10.7554/eLife.46440. PMID: 31038124; PMCID: PMC6491035.
- Sacristán S, Goss EM, Eves-van den Akker S. How Do Pathogens Evolve Novel Virulence Activities? Mol Plant Microbe Interact. 2021 Jun;34(6):576-586. doi: 10.1094/MPMI-09-20-0258-IA. Epub 2021 Jul 23. PMID: 33522842.
- Seal S, Dharmarajan G, Khan I. Evolution of pathogen tolerance and emerging infections: A missing experimental paradigm. Elife. 2021 Sep 21;10:e68874. doi: 10.7554/eLife.68874. PMID: 34544548; PMCID: PMC8455132.
- Li W, Shi Z, Yu M, Ren W, Smith C, Epstein JH, Wang H, Crameri G, Hu Z, Zhang H, Zhang J, McEachern J, Field H, Daszak P, Eaton BT, Zhang S, Wang LF. Bats are natural reservoirs of SARS-like coronaviruses. Science. 2005 Oct 28;310(5748):676-9. doi: 10.1126/science.1118391. Epub 2005 Sep 29. PMID: 16195424.
- Wang LF, Shi Z, Zhang S, Field H, Daszak P, Eaton BT. Review of bats and SARS. Emerg Infect Dis. 2006 Dec;12(12):1834-40. doi: 10.3201/eid1212.060401. PMID: 17326933; PMCID: PMC3291347.
- Sharp PM, Hahn BH. Origins of HIV and the AIDS pandemic. Cold Spring Harb Perspect Med. 2011 Sep;1(1):a006841. doi: 10.1101/cshperspect.a006841. PMID: 22229120; PMCID: PMC3234451.
- Marí Saéz A, Weiss S, Nowak K, Lapeyre V, Zimmermann F, Düx A, Kühl HS, Kaba M, Regnaut S, Merkel K, Sachse A, Thiesen U, Villányi L, Boesch C, Dabrowski PW, Radonić A, Nitsche A, Leendertz SA, Petterson S, Becker S, Krähling V, Couacy-Hymann E, Akoua-Koffi C, Weber N, Schaade L, Fahr J, Borchert M, Gogarten JF, Calvignac-Spencer S, Leendertz FH. Investigating the zoonotic origin of the West African Ebola epidemic. EMBO Mol Med. 2015 Jan;7(1):17-23. doi: 10.15252/emmm.201404792. PMID: 25550396; PMCID: PMC4309665.
- Keesing F, Belden LK, Daszak P, Dobson A, Harvell CD, Holt RD, Hudson P, Jolles A, Jones KE, Mitchell CE, Myers SS, Bogich T, Ostfeld RS. Impacts of biodiversity on the emergence and transmission of infectious diseases. Nature. 2010 Dec 2;468(7324):647-52. doi: 10.1038/nature09575. PMID: 21124449; PMCID: PMC7094913.
- De Vos A, Cumming GS, Cumming D, Ament JM, Baum J, Clements H, Grewar J, Maciejewski K, and Moore C. Pathogens, disease, and the social-ecological resilience of protected areas. Ecology and Society. 2016; 21:20. http://dx.doi.org/10.5751/ ES-07984-210120
- Albery GF, Becker DJ, Brierley L, Brook CE, Christofferson RC, Cohen LE, Dallas TA, Eskew EA, Fagre A, Farrell MJ, Glennon E, Guth S, Joseph MB, Mollentze N, Neely BA, Poisot T, Rasmussen AL, Ryan SJ, Seifert S, Sjodin AR, Sorrell EM, Carlson CJ. The science of the host-virus network. Nat Microbiol. 2021 Dec;6(12):1483-1492. doi: 10.1038/s41564-021-00999-5. Epub 2021 Nov 24. PMID: 34819645.
- Keesing F, Ostfeld RS. Impacts of biodiversity and biodiversity loss on zoonotic diseases. Proc Natl Acad Sci U S A. 2021 Apr 27;118(17):e2023540118. doi: 10.1073/pnas.2023540118. PMID: 33820825; PMCID: PMC8092607.
- Wells K, Flynn R. Managing host-parasite interactions in humans and wildlife in times of global change. Parasitol Res. 2022 Nov;121(11):3063-3071. doi: 10.1007/s00436-022-07649-7. Epub 2022 Sep 6. PMID: 36066742; PMCID: PMC9446624.
- Johnson CK, Hitchens PL, Pandit PS, Rushmore J, Evans TS, Young CCW, Doyle MM. Global shifts in mammalian population trends reveal key predictors of virus spillover risk. Proc Biol Sci. 2020 Apr 8;287(1924):20192736. doi: 10.1098/rspb.2019.2736. Epub 2020 Apr 8. PMID: 32259475; PMCID: PMC7209068.
- Karesh WB, Cook RA, Bennett EL, Newcomb J. Wildlife trade and global disease emergence. Emerg Infect Dis. 2005 Jul;11(7):1000-2. doi: 10.3201/eid1107.050194. PMID: 16022772; PMCID: PMC3371803.
- Wolfe ND, Daszak P, Kilpatrick AM, Burke DS. Bushmeat hunting, deforestation, and prediction of zoonoses emergence. Emerg Infect Dis. 2005 Dec;11(12):1822-7. doi: 10.3201/eid1112.040789. PMID: 16485465; PMCID: PMC3367616.
- Karesh WB, Noble E. The bushmeat trade: increased opportunities for transmission of zoonotic disease. Mt Sinai J Med. 2009 Oct;76(5):429-34. doi: 10.1002/msj.20139. PMID: 19787649.
- Kreuder Johnson C, Hitchens PL, Smiley Evans T, Goldstein T, Thomas K, Clements A, Joly DO, Wolfe ND, Daszak P, Karesh WB, Mazet JK. Spillover and pandemic properties of zoonotic viruses with high host plasticity. Sci Rep. 2015 Oct 7;5:14830. doi: 10.1038/srep14830. PMID: 26445169; PMCID: PMC4595845.
- Hime JM, Keymer IF, Baxter CJ. Measles in recently imported colobus monkeys (Colobus guereza). Vet Rec. 1975 Nov 15;97(20):392. doi: 10.1136/vr.97.20.392-a. PMID: 813359.
- Bermejo M, Rodríguez-Teijeiro JD, Illera G, Barroso A, Vilà C, Walsh PD. Ebola outbreak killed 5000 gorillas. Science. 2006 Dec 8;314(5805):1564. doi: 10.1126/science.1133105. PMID: 17158318.
- Caillaud D, Levréro F, Cristescu R, Gatti S, Dewas M, Douadi M, Gautier-Hion A, Raymond M, Ménard N. Gorilla susceptibility to Ebola virus: the cost of sociality. Curr Biol. 2006 Jul 11;16(13):R489-91. doi: 10.1016/j.cub.2006.06.017. PMID: 16824905.
- Genton C, Cristescu R, Gatti S, Levréro F, Bigot E, Caillaud D, Pierre JS, Ménard N. Recovery potential of a western lowland gorilla population following a major Ebola outbreak: results from a ten year study. PLoS One. 2012;7(5):e37106. doi: 10.1371/journal.pone.0037106. Epub 2012 May 23. PMID: 22649511; PMCID: PMC3359368.
- Zhou H, Ji J, Chen X, Bi Y, Li J, Wang Q, Hu T, Song H, Zhao R, Chen Y, Cui M, Zhang Y, Hughes AC, Holmes EC, Shi W. Identification of novel bat coronaviruses sheds light on the evolutionary origins of SARS-CoV-2 and related viruses. Cell. 2021 Aug 19;184(17):4380-4391.e14. doi: 10.1016/j.cell.2021.06.008. Epub 2021 Jun 9. PMID: 34147139; PMCID: PMC8188299.
- Delaune D, Hul V, Karlsson EA, Hassanin A, Ou TP, Baidaliuk A, Gámbaro F, Prot M, Tu VT, Chea S, Keatts L, Mazet J, Johnson CK, Buchy P, Dussart P, Goldstein T, Simon-Lorière E, Duong V. A novel SARS-CoV-2 related coronavirus in bats from Cambodia. Nat Commun. 2021 Nov 9;12(1):6563. doi: 10.1038/s41467-021-26809-4. PMID: 34753934; PMCID: PMC8578604.
- Wacharapluesadee S, Tan CW, Maneeorn P, Duengkae P, Zhu F, Joyjinda Y, Kaewpom T, Chia WN, Ampoot W, Lim BL, Worachotsueptrakun K, Chen VC, Sirichan N, Ruchisrisarod C, Rodpan A, Noradechanon K, Phaichana T, Jantarat N, Thongnumchaima B, Tu C, Crameri G, Stokes MM, Hemachudha T, Wang LF. Evidence for SARS-CoV-2 related coronaviruses circulating in bats and pangolins in Southeast Asia. Nat Commun. 2021 Feb 9;12(1):972. doi: 10.1038/s41467-021-21240-1. Erratum in: Nat Commun. 2021 Feb 25;12(1):1430. PMID: 33563978; PMCID: PMC7873279.
- Murakami S, Kitamura T, Suzuki J, Sato R, Aoi T, Fujii M, Matsugo H, Kamiki H, Ishida H, Takenaka-Uema A, Shimojima M, Horimoto T. Detection and Characterization of Bat Sarbecovirus Phylogenetically Related to SARS-CoV-2, Japan. Emerg Infect Dis. 2020 Dec;26(12):3025-3029. doi: 10.3201/eid2612.203386. PMID: 33219796; PMCID: PMC7706965.
- Rahalkar MC, Bahulikar RA. Lethal Pneumonia Cases in Mojiang Miners (2012) and the Mineshaft Could Provide Important Clues to the Origin of SARS-CoV-2. Front Public Health. 2020 Oct 20;8:581569. doi: 10.3389/fpubh.2020.581569. PMID: 33194988; PMCID: PMC7606707.
- Liu P, Jiang JZ, Wan XF, Hua Y, Li L, Zhou J, Wang X, Hou F, Chen J, Zou J, Chen J. Are pangolins the intermediate host of the 2019 novel coronavirus (SARS-CoV-2)? PLoS Pathog. 2020 May 14;16(5):e1008421. doi: 10.1371/journal.ppat.1008421. Erratum in: PLoS Pathog. 2021 Jun 9;17(6):e1009664. PMID: 32407364; PMCID: PMC7224457.
- Xiao K, Zhai J, Feng Y, Zhou N, Zhang X, Zou JJ, Li N, Guo Y, Li X, Shen X, Zhang Z, Shu F, Huang W, Li Y, Zhang Z, Chen RA, Wu YJ, Peng SM, Huang M, Xie WJ, Cai QH, Hou FH, Chen W, Xiao L, Shen Y. Isolation of SARS-CoV-2-related coronavirus from Malayan pangolins. Nature. 2020 Jul;583(7815):286-289. doi: 10.1038/s41586-020-2313-x. Epub 2020 May 7. Erratum in: Nature. 2021 Dec;600(7887):E8-E10. PMID: 32380510.
- Wahba L, Jain N, Fire AZ, Shoura MJ, Artiles KL, McCoy MJ, Jeong DE. An Extensive Meta-Metagenomic Search Identifies SARS-CoV-2-Homologous Sequences in Pangolin Lung Viromes. mSphere. 2020 May 6;5(3):e00160-20. doi: 10.1128/mSphere.00160-20. PMID: 32376697; PMCID: PMC7203451.
- Temmam S, Vongphayloth K, Baquero E, Munier S, Bonomi M, Regnault B, Douangboubpha B, Karami Y, Chrétien D, Sanamxay D, Xayaphet V, Paphaphanh P, Lacoste V, Somlor S, Lakeomany K, Phommavanh N, Pérot P, Dehan O, Amara F, Donati F, Bigot T, Nilges M, Rey FA, van der Werf S, Brey PT, Eloit M. Bat coronaviruses related to SARS-CoV-2 and infectious for human cells. Nature. 2022 Apr;604(7905):330-336. doi: 10.1038/s41586-022-04532-4. Epub 2022 Feb 16. Erratum in: Nature. 2022 Jul;607(7920):E19. PMID: 35172323.
- Jackson B, Boni MF, Bull MJ, Colleran A, Colquhoun RM, Darby AC, Haldenby S, Hill V, Lucaci A, McCrone JT, Nicholls SM, O'Toole Á, Pacchiarini N, Poplawski R, Scher E, Todd F, Webster HJ, Whitehead M, Wierzbicki C; COVID-19 Genomics UK (COG-UK) Consortium; Loman NJ, Connor TR, Robertson DL, Pybus OG, Rambaut A. Generation and transmission of interlineage recombinants in the SARS-CoV-2 pandemic. Cell. 2021 Sep 30;184(20):5179-5188.e8. doi: 10.1016/j.cell.2021.08.014. Epub 2021 Aug 17. PMID: 34499854; PMCID: PMC8367733.
- Rochman ND, Wolf YI, Faure G, Mutz P, Zhang F, Koonin EV. Ongoing global and regional adaptive evolution of SARS-CoV-2. Proc Natl Acad Sci U S A. 2021 Jul 20;118(29):e2104241118. doi: 10.1073/pnas.2104241118. Epub 2021 Jul 2. PMID: 34292871; PMCID: PMC8307621.
- Our World in Data: Coronavirus (COVID-19) Deaths. Feb 18, 2023. https://ourworldindata.org/covid-deaths
- Pickering B, Lung O, Maguire F, Kruczkiewicz P, Kotwa JD, Buchanan T, Gagnier M, Guthrie JL, Jardine CM, Marchand-Austin A, Massé A, McClinchey H, Nirmalarajah K, Aftanas P, Blais-Savoie J, Chee HY, Chien E, Yim W, Banete A, Griffin BD, Yip L, Goolia M, Suderman M, Pinette M, Smith G, Sullivan D, Rudar J, Vernygora O, Adey E, Nebroski M, Goyette G, Finzi A, Laroche G, Ariana A, Vahkal B, Côté M, McGeer AJ, Nituch L, Mubareka S, Bowman J. Divergent SARS-CoV-2 variant emerges in white-tailed deer with deer-to-human transmission. Nat Microbiol. 2022 Dec;7(12):2011-2024. doi: 10.1038/s41564-022-01268-9. Epub 2022 Nov 10. Erratum in: Nat Microbiol. 2022 Dec 12;: PMID: 36357713; PMCID: PMC9712111.
- Ribeiro JP, Barbosa-Povoa A. Supply Chain Resilience: Definitions and Quantitative Modelling Approaches-A literature review. Comput. Ind. Eng. 2018; 115:109-122
- Lenzen M, Li M, Malik A, Pomponi F, Sun YY, Wiedmann T, Faturay F, Fry J, Gallego B, Geschke A, Gómez-Paredes J, Kanemoto K, Kenway S, Nansai K, Prokopenko M, Wakiyama T, Wang Y, Yousefzadeh M. Global socio-economic losses and environmental gains from the Coronavirus pandemic. PLoS One. 2020 Jul 9;15(7):e0235654. doi: 10.1371/journal.pone.0235654. PMID: 32645023; PMCID: PMC7347123.
- Soyyiğit S, Eren E. Global supply and demand of medical goods in the fight against Covid-19: a network analysis. Asia-Pacific Journal of Regional Science. 2022; 6: 1221–1247
- Riaz MMA, Ahmad U, Mohan A, Dos Santos Costa AC, Khan H, Babar MS, Hasan MM, Essar MY, Zil-E-Ali A. Global impact of vaccine nationalism during COVID-19 pandemic. Trop Med Health. 2021 Dec 29;49(1):101. doi: 10.1186/s41182-021-00394-0. PMID: 34963494; PMCID: PMC8714455.
- Bortolotti L, Murphy-Hollies K. Exceptionalism at the time of covid-19: Where nationalism meets irrationality. Danish Yearbook of Philosophy. 2022; 55: 90-111
- Mylonas H, Whalley N. Pandemic Nationalism. Nationalities Papers. 2022; 50: 3–12
- Finch WH, Hernández Finch ME. Poverty and Covid-19: Rates of Incidence and Deaths in the United States During the First 10 Weeks of the Pandemic. Front Sociol. 2020 Jun 15;5:47. doi: 10.3389/fsoc.2020.00047. PMID: 33869454; PMCID: PMC8022686.
- Davies JB. Economic Inequality and COVID-19 Deaths and Cases in the First Wave: A Cross-Country Analysis. Can Public Policy. 2021 Dec 1;47(4):537-553. doi: 10.3138/cpp.2021-033. PMID: 36039094; PMCID: PMC9395158.
- Liao TF, De Maio F. Association of Social and Economic Inequality With Coronavirus Disease 2019 Incidence and Mortality Across US Counties. JAMA Netw Open. 2021 Jan 4;4(1):e2034578. doi: 10.1001/jamanetworkopen.2020.34578. Erratum in: JAMA Netw Open. 2021 May 3;4(5):e2114136. PMID: 33471120; PMCID: PMC7818127.
- Perry BL, Aronson B, Pescosolido BA. Pandemic precarity: COVID-19 is exposing and exacerbating inequalities in the American heartland. Proc Natl Acad Sci U S A. 2021 Feb 23;118(8):e2020685118. doi: 10.1073/pnas.2020685118. PMID: 33547252; PMCID: PMC7923675.
- Dauderstädt M. International Inequality and the COVID-19 Pandemic. Inter Econ. 2022;57(1):40-46. doi: 10.1007/s10272-022-1026-9. Epub 2022 Feb 12. PMID: 35194239; PMCID: PMC8853295.
Article Alerts
Subscribe to our articles alerts and stay tuned.
 This work is licensed under a Creative Commons Attribution 4.0 International License.
This work is licensed under a Creative Commons Attribution 4.0 International License.



 Save to Mendeley
Save to Mendeley
